Running ML inference through a Huey Task Queue and FastAPI
Learn how to train a simple CNN in PyTorch and how to convert it to ONNX for deployment.
August 26, 2024
07m Read
By: Abhilaksh Singh Reen
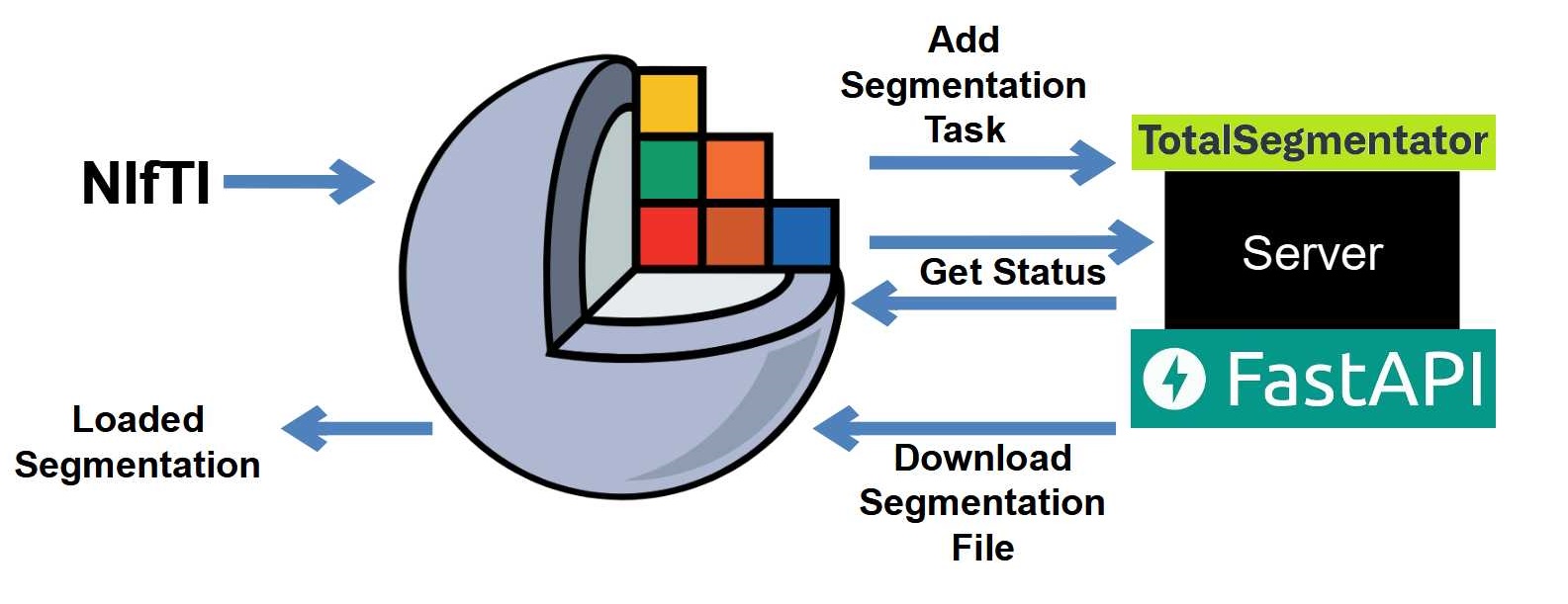
In today's Post, we'll build a REST API that allows us to segment CT volumes using TotalSegmentator. Our system will involve two applications connected through the Huey task queue: a server that a client-side application can interact with to upload volumes and queue segmentation tasks and a worker application that consumes these tasks to segment the uploaded volumes and save the results.
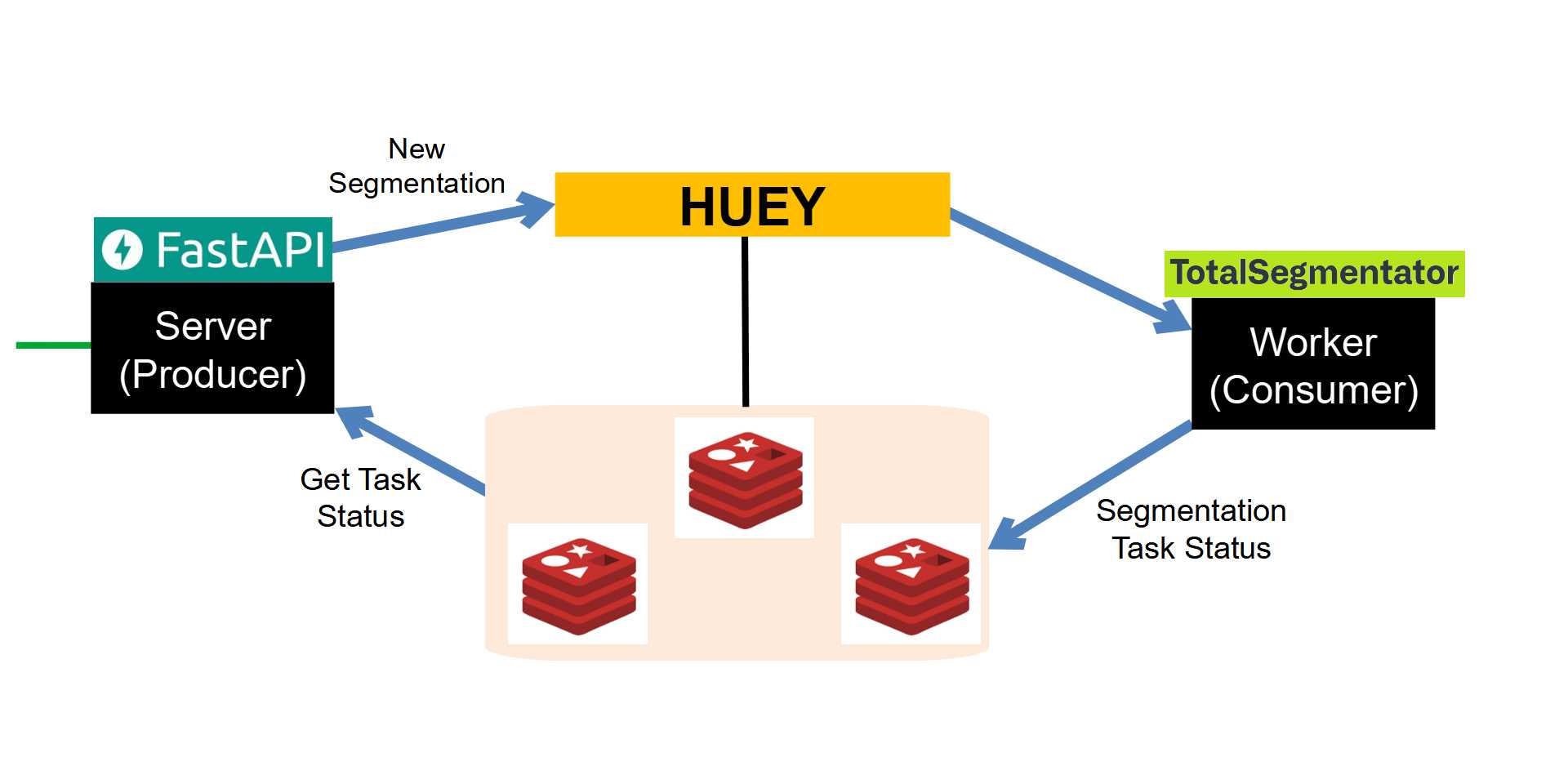
Here's an image describing the System Architecture:

You can find the code for this project in this GitHub Repository.
Server and Worker
Interacting with Redis
We'll be using Redis for two purposes:
1) As the Huey task broker.
1) For storing the statuses of our tasks.
Redis Server
We can run a lightweight Redis Docker container for development purposes
docker run -d --name redis-dev -p 6379:6379 redis:7.2.4-alpineA list of Redis Docker images can be found here.
Then, check if the Redis container started.
docker psYou should see a container with the name redis-dev up and running.
Redis Client
Inside our project's directory, let's create a folder called server-worker, and inside it create another folder called src. Inside this folder, we create a file called redis_client.py.
from os import environ
from redis import Redis
redis_host = environ["REDIS_HOST"]
redis_port = environ["REDIS_PORT"]
redis_db = environ["REDIS_DB"]
redis_password = environ.get("REDIS_PASSWORD", None)
redis_client = Redis(host=redis_host, port=redis_port, db=redis_db, password=redis_password, decode_responses=True)We can create an empty file __init__.py in the server-worker/src folder to turn this into an importable package.
Tasks
Inside the same folder, let's create a file called dirs.py to store some directory paths for our convenience.
from os import makedirs
from os.path import dirname, join as path_join
src_dir = dirname(__file__)
server_worker_dir = dirname(src_dir)
project_dir = dirname(server_worker_dir)
data_dir = path_join(project_dir, "data")
makedirs(data_dir, exist_ok=True)Now, let's create a file called tasks.py in this directory. This will store out Huey tasks.
from os import environ
from os.path import join as path_join
from subprocess import run as subprocess_run, CalledProcessError
from time import sleep
from huey import RedisHuey
from src.redis_client import redis_client
from src.dirs import data_dir
redis_host = environ["REDIS_HOST"]
task_delay = float(environ.get("TASK_DELAY", 0))
huey = RedisHuey('entrypoint', host=redis_host)
task_status_map_name = "segmentation_task_status"
@huey.task()
def segment_volume(task_id):
volume_path = path_join(data_dir, task_id, "volume.nii.gz")
segmentation_path = path_join(data_dir, task_id, "segmentation.nii.gz")
redis_client.hset(task_status_map_name, task_id, "processing")
run_total_segmentator_command = f"TotalSegmentator -i {volume_path} -o {segmentation_path} --ml"
try:
_result = subprocess_run(run_total_segmentator_command, shell=True, check=True)
redis_client.hset(task_status_map_name, task_id, "completed")
except CalledProcessError as _e:
print(f"Failed to segment volume for task_id {task_id}.")
print(f" segmentation command: {run_total_segmentator_command}")
redis_client.hset(task_status_map_name, task_id, "failed")The Redis hashmap with the name given by task_status_map_name will be used to store the statuses of our tasks. When we first receive a task (inside the segment_volume function), we update its status to processing. Then, we run TotalSegmentator on the volume using subprocess.run. If the process runs successfully, we update the status of the task to completed. The segmentation will be saved at the location specified by segmentation_path. If the process fails with an error, we mark the task as failed.
FastAPI Server
Inside the server-worker/src directory, let's create a file called app.py, which will contain our FastAPI application. As of now, we have three needs from this application:
1) Serve the static files present in the data folder (in production this should be handled by your web server).
2) An endpoint where we can upload a Nifti file to start the segmentation task.
3) An endpoint where we can get the status/result of a task.
Here is the app.py file:
from os import makedirs
from os.path import basename as path_basename, join as path_join
from uuid import uuid4
from fastapi import FastAPI, UploadFile, File
from fastapi.responses import JSONResponse
from fastapi.staticfiles import StaticFiles
from src.dirs import data_dir
from src.redis_client import redis_client
from src.tasks import segment_volume, task_status_map_name
app = FastAPI()
app.mount("/data", StaticFiles(directory=data_dir), name="data_dir")
@app.post("/api/add-segmentation-task")
async def upload_file(file: UploadFile = File(...)):
task_id = str(uuid4())
task_dir = path_join(data_dir, task_id)
makedirs(task_dir)
volume_save_path = path_join(task_dir, "volume.nii.gz")
# Save the file to the disk
with open(volume_save_path, "wb") as buffer:
while True:
chunk = await file.read(10000)
if not chunk:
break
buffer.write(chunk)
# Queue the task
redis_client.hset(task_status_map_name, task_id, "queued")
segment_volume(task_id)
return JSONResponse(content={
'success': True,
'result': {
'taskId': task_id,
},
}, status_code=206)
@app.get("/api/get-segmentation-task-result")
async def get_segmentation(task_id: str):
task_status = redis_client.hget(task_status_map_name, task_id)
response_content = {
'success': True,
'result': {
'taskId': task_id,
'status': task_status,
'volumeFileUrl': path_join(path_basename(data_dir), task_id, "volume.nii.gz"),
},
}
if task_status == "completed":
response_content['result']['segmentationFileUrl'] = path_join(path_basename(data_dir), task_id, "segmentation.nii.gz")
return JSONResponse(content=response_content, status_code=200)Running the App
Make sure you have a Redis server running.
We'll need to create two environments: one for the server and one for the worker. I'll be using venv for creating and managing the environments.
Server
python3 -m venv env_server
source env_server/bin/activateNext, we install the following packages:
pip install fastapi pydantic uvicorn python-multipart redis hueyWe can now run the server from the server-worker directory using:
REDIS_HOST=localhost REDIS_PORT=6379 REDIS_DB=0 uvicorn src.app:app --port=8000 --host=0.0.0.0Worker
python3 -m venv env_worker
source env_worker/bin/activateFor the worker, we will also install the TotalSegmentator apart from the packages we installed for the server.
pip install fastapi pydantic uvicorn python-multipart redis huey TotalSegmentatorFrom the server-worker directory, run the following command to start the worker process.
REDIS_HOST=localhost REDIS_PORT=6379 REDIS_DB=0 huey_consumer.py src.tasks.hueyTests
With both the server and worker applications running, we can make a test case for our segmentation process in which we will perform the following operations:
1) Add a segmentation task by uploading a sample volume.
1) Repeatedly query the status endpoint and wait for the task to complete (or fail).
1) Download the volume and segmentation files.
1) Read the volume and segmentation files and verify if they have the same shape.
Inside the project directory, create a new directory called tests. This should be on the same level as the server-worker folder. Here, we'll have a dirs.py file quite similar to the dirs.py in server-worker/src.
from os import makedirs
from os.path import dirname, join as path_join
tests_dir = dirname(__file__)
project_dir = dirname(tests_dir)
data_dir = path_join(project_dir, "data")
makedirs(data_dir, exist_ok=True)Next, create a file called utils.py to contain a couple of utility functions:
from sys import stdout
import requests
def print_inline(string):
print(string, end="")
stdout.flush()
def download_file(url, file_path):
response = requests.get(url)
if response.status_code != 200:
print(f"Failed to download file. Response status code: {response.status_code}")
return False
with open(file_path, "wb") as f:
f.write(response.content)
return TrueNow, let's write our core testing logic. Create a file called test_apis.py and add the following lines
from os import makedirs
from os.path import basename as path_basename, join as path_join
from shutil import rmtree
from time import sleep, time
import unittest
from nibabel import load as nib_load
import requests
from dirs import data_dir, tests_dir
from utils import download_file, print_inline
class TestAPIs(unittest.TestCase):
base_url = "http://localhost:8000"
api_base_url = "http://localhost:8000/api"
test_volume_path = path_join(data_dir, "testing", "volume.nii.gz")
def test_add_segmentation_task_and_get_result(self):
pass
if __name__ == "__main__":
unittest.main()The test_add_segmentation_task_and_get_result function will contain the logic for performing the four aforementioned testing operations. Inside the data folder in the project's root directory, we'll create another folder called testing and place a CT Nifti image called volume.nii.gz in it. Let's upload this volume for segmentation in our test.
add_segmentation_task_endpoint = f"{self.api_base_url}/add-segmentation-task"
print_inline("Adding task ...")
with open(self.test_volume_path, "rb") as f:
files = {"file": f}
add_response = requests.post(add_segmentation_task_endpoint, files=files)
self.assertEqual(add_response.status_code, 206)
self.assertEqual(add_response.headers['content-type'], "application/json")
add_response_data = add_response.json()
self.assertEqual(add_response_data.get('success', None), True)
self.assertNotEqual(add_response_data.get('result', {}).get('taskId', None), None)
print(" ok.")Then, we will repeatedly fetch the status of the task until it is completed or failed. If the status is failed, we fail the test. If the status is completed, we make sure that the segmentationFileUrl in the response is actually what is expected for the task_id.
task_id = add_response_data['result']['taskId']
get_segmentation_task_result_endpoint = f"{self.api_base_url}/get-segmentation-task-result?task_id={task_id}"
expected_volume_file_url = path_join(path_basename(data_dir), task_id, "volume.nii.gz")
expected_segmentation_file_url = path_join(path_basename(data_dir), task_id, "segmentation.nii.gz")
print_inline("Getting task status ")
while True:
get_result_response = requests.get(get_segmentation_task_result_endpoint)
self.assertEqual(get_result_response.status_code, 200)
self.assertEqual(get_result_response.headers['content-type'], "application/json")
get_result_response_data = get_result_response.json()
self.assertEqual(get_result_response_data.get('success', None), True)
self.assertNotEqual(get_result_response_data.get('result', {}).get('status', None), None)
self.assertEqual(get_result_response_data.get('result', {}).get('taskId', None), task_id)
self.assertEqual(
get_result_response_data.get('result', {}).get('volumeFileUrl', None),
expected_volume_file_url
)
print_inline(".")
if get_result_response_data['result']['status'] == "failed":
self.fail("Server failed to segment the uploaded volume.")
break
if get_result_response_data['result']['status'] == "completed":
self.assertEqual(
get_result_response_data.get('result', {}).get('segmentationFileUrl', None).strip(),
expected_segmentation_file_url.strip()
)
break
sleep(1)
print(" ok.")Next, we download the volume and segmentation files.
print_inline("Downloading files ... ")
volume_file_url = f"{self.base_url}/{expected_volume_file_url}"
segmentation_file_url = f"{self.base_url}/{expected_segmentation_file_url}"
temp_dir = path_join(tests_dir, "temp")
makedirs(temp_dir, exist_ok=True)
volume_file_path = path_join(temp_dir, f"volume-{time()}.nii.gz")
segmentation_file_path = path_join(temp_dir, f"segmentation-{time()}.nii.gz")
if not download_file(volume_file_url, volume_file_path):
self.fail("Failed to download volume file from server.")
print_inline(" volume download ok ... ")
if not download_file(segmentation_file_url, segmentation_file_path):
self.fail("Failed to download segmentation file from server.")
print_inline(" segmentation download ok ... ")Finally, we check whether both the files have the same shape and then delete the downloaded files.
volume_file_data = nib_load(volume_file_path).get_fdata()
segmentation_file_data = nib_load(volume_file_path).get_fdata()
self.assertTupleEqual(volume_file_data.shape, segmentation_file_data.shape)
print(" shapes ok.")
rmtree(temp_dir)Here's the entire test_apis.py file
from os import makedirs
from os.path import basename as path_basename, join as path_join
from shutil import rmtree
from time import sleep, time
import unittest
from nibabel import load as nib_load
import requests
from dirs import data_dir, tests_dir
from utils import download_file, print_inline
class TestAPIs(unittest.TestCase):
base_url = "http://localhost:8000"
api_base_url = "http://localhost:8000/api"
test_volume_path = path_join(data_dir, "testing", "volume.nii.gz")
def test_add_segmentation_task_and_get_result(self):
add_segmentation_task_endpoint = f"{self.api_base_url}/add-segmentation-task"
print_inline("Adding task ...")
with open(self.test_volume_path, "rb") as f:
files = {"file": f}
add_response = requests.post(add_segmentation_task_endpoint, files=files)
self.assertEqual(add_response.status_code, 206)
self.assertEqual(add_response.headers['content-type'], "application/json")
add_response_data = add_response.json()
self.assertEqual(add_response_data.get('success', None), True)
self.assertNotEqual(add_response_data.get('result', {}).get('taskId', None), None)
print(" ok.")
task_id = add_response_data['result']['taskId']
get_segmentation_task_result_endpoint = f"{self.api_base_url}/get-segmentation-task-result?task_id={task_id}"
expected_volume_file_url = path_join(path_basename(data_dir), task_id, "volume.nii.gz")
expected_segmentation_file_url = path_join(path_basename(data_dir), task_id, "segmentation.nii.gz")
print_inline("Getting task status ")
while True:
get_result_response = requests.get(get_segmentation_task_result_endpoint)
self.assertEqual(get_result_response.status_code, 200)
self.assertEqual(get_result_response.headers['content-type'], "application/json")
get_result_response_data = get_result_response.json()
self.assertEqual(get_result_response_data.get('success', None), True)
self.assertNotEqual(get_result_response_data.get('result', {}).get('status', None), None)
self.assertEqual(get_result_response_data.get('result', {}).get('taskId', None), task_id)
self.assertEqual(
get_result_response_data.get('result', {}).get('volumeFileUrl', None),
expected_volume_file_url
)
print_inline(".")
if get_result_response_data['result']['status'] == "failed":
self.fail("Server failed to segment the uploaded volume.")
break
if get_result_response_data['result']['status'] == "completed":
self.assertEqual(
get_result_response_data.get('result', {}).get('segmentationFileUrl', None).strip(),
expected_segmentation_file_url.strip()
)
break
sleep(1)
print(" ok.")
print_inline("Downloading files ... ")
volume_file_url = f"{self.base_url}/{expected_volume_file_url}"
segmentation_file_url = f"{self.base_url}/{expected_segmentation_file_url}"
temp_dir = path_join(tests_dir, "temp")
makedirs(temp_dir, exist_ok=True)
volume_file_path = path_join(temp_dir, f"volume-{time()}.nii.gz")
segmentation_file_path = path_join(temp_dir, f"segmentation-{time()}.nii.gz")
if not download_file(volume_file_url, volume_file_path):
self.fail("Failed to download volume file from server.")
print_inline(" volume download ok ... ")
if not download_file(segmentation_file_url, segmentation_file_path):
self.fail("Failed to download segmentation file from server.")
print_inline(" segmentation download ok ... ")
volume_file_data = nib_load(volume_file_path).get_fdata()
segmentation_file_data = nib_load(volume_file_path).get_fdata()
self.assertTupleEqual(volume_file_data.shape, segmentation_file_data.shape)
print(" shapes ok.")
rmtree(temp_dir)
if __name__ == "__main__":
unittest.main()Let's create a new environment for the tests
python3 -m venv env_test
source env_test/bin/activateAnd, install the following packages
pip install requests nibabelWe can run the tests by running the following command from the tests directory:
python3 test_apis.pyYou should see an output saying that one test ran successfully.
Conclusion
We have successfully created and tested the APIs that allow us to convert a CT scan volume into a segmentation using TotalSegmentator.
3D Slicer is a tool that can be used to visualize the segmentations we generated today. If you'd like to know more about 3D Slicer and how to customize it to fit some of your own requirements, you can check out this series where I cover how to build a Slicer Extension or specifically this part, in which, we make a Slicer Extension to work with the API we created today.
See you next time :)